Dartmouth Kidney Cancer Histology Dataset
This dataset comprises 563 hematoxylin and eosin (H&E)-stained formalin-fixed paraffin-embedded (FFPE) whole-slide images of renal cell carcinoma (RCC) from the Department of Pathology and Laboratory Medicine at Dartmouth-Hitchcock Medical Center (DHMC). The dataset is de-identified and released with permission from Dartmouth-Hitchcock Health (D-HH) Institutional Review Board (IRB). All whole-slide images are labeled for the predominant pattern of renal cell carcinoma according to the consensus opinion of two pathologists, Drs. Bing Ren and Ryland Richards, at the Department of Pathology and Laboratory Medicine at DHMC. For more information about this dataset, please refer to “Development and Evaluation of a Deep Neural Network for Histologic Classification of Renal Cell Carcinoma on Biopsy and Surgical Resection Slides”.
Renal Cell Carcinoma Subtypes Classification
Histological classification of RCC subtypes has significant implication in the prognosis and treatment of patients. However, this task is challenging due to the varied appearances and mixed morphologic features of renal tumors.
This dataset and its associated classification labels aim to foster collaboration with the research community and facilitate developing and evaluating new methodologies for accurate histology image analysis in this domain. The classification labels in our dataset indicate the predominant histological pattern of each whole-slide image and are as follows:
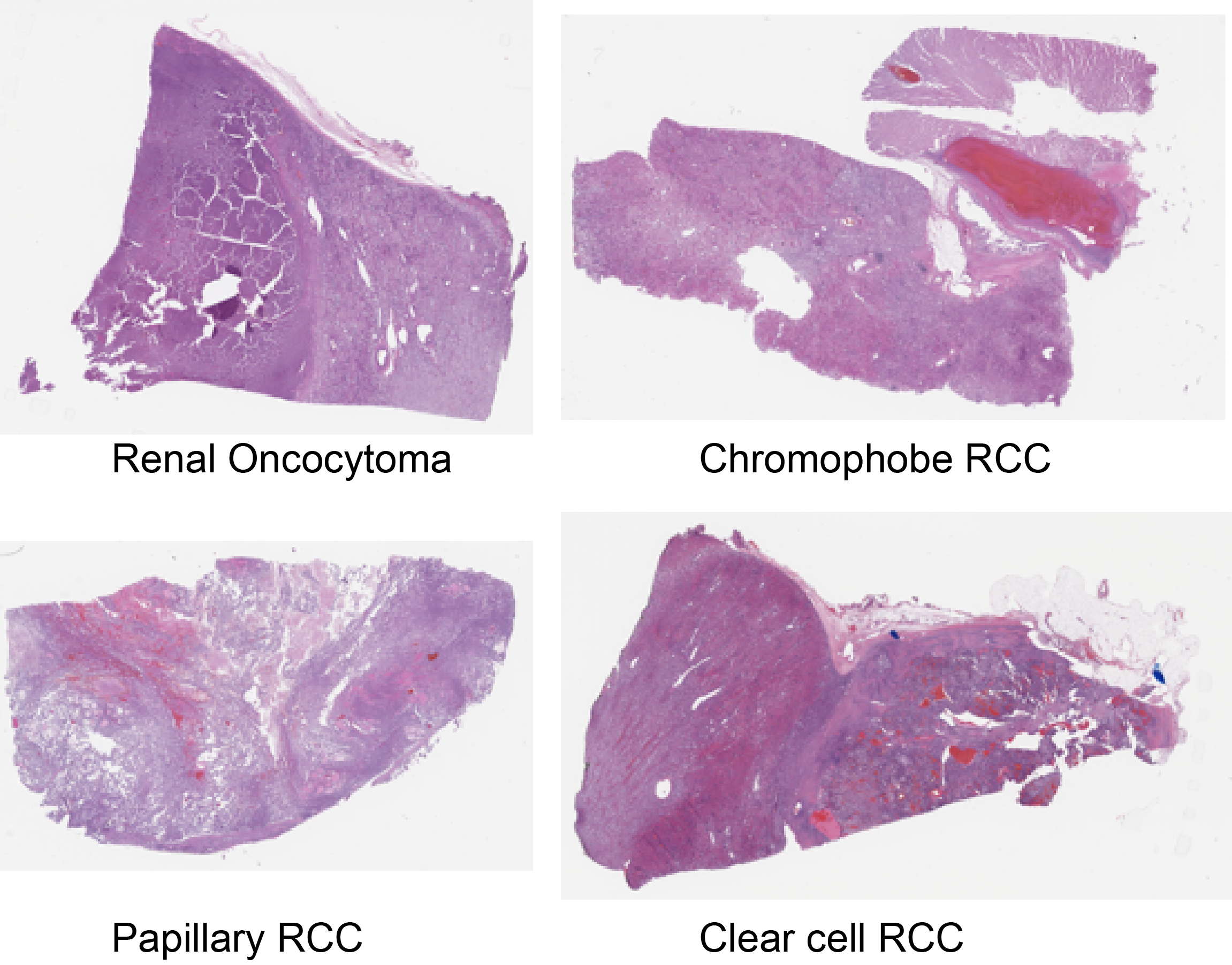
- Renal Oncocytoma
- Chromophobe RCC
- Clear cell RCC
- Papillary RCC
Sample Whole-Slide Images
Dataset Description
The dataset includes:
- DHMC_wsi_01.zip - (Resection Slides: 1-49, 7.3 GB)
- DHMC_wsi_02.zip - (Resection Slides: 50-99, 8.4 GB)
- DHMC_wsi_03.zip - (Resection Slides: 100-149, 9.3 GB)
- DHMC_wsi_04.zip - (Resection Slides: 150-199, 11.0 GB)
- DHMC_wsi_05.zip - (Resection Slides: 200-249, 8.7 GB)
- DHMC_wsi_06.zip - (Resection Slides: 250-299, 9.2 GB)
- DHMC_wsi_07.zip - (Resection Slides: 300-349, 8.9 GB)
- DHMC_wsi_08.zip - (Resection Slides: 350-399, 8.4 GB)
- DHMC_wsi_09.zip - (Resection Slides: 400-449, 7.8 GB)
- DHMC_wsi_10.zip - (Resection Slides: 450-484, 5.8 GB)
- DHMC_wsi_11.zip - (Biopsy Slides: 485-563, 4.7 GB)
- MetaData.csv
Each zip file contains whole-slide images in .png image format, which were originally scanned by an Aperio AT2 whole-slide scanner at 20x magnification and converted to Portable Network Graphics, or PNG format using libvips at 5x magnification. The list of scanned slides, as well as their classes, slide types, and a data split (i.e., train/validation/test split that are used in the published work), are available in MetaData.csv. In addition to surgical resection slides, DHMC_wsi_11.zip stores biopsy slides that were used as an extended test set in our work.
Code Repository
DeepSlide, our open-source framework for histology image analysis in PyTorch, is available to develop deep learning models for whole-slide image classification.
Accessing Dataset
Please fill out the form below to receive the links to download the dataset by email.
Citation
If you use this dataset, please cite the corresponding paper:
Mengdan Zhu, Bing Ren, Ryland Richards, Matthew Suriawinata, Naofumi Tomita, Saeed Hassanpour, "Development and Evaluation of a Deep Neural Network for Histologic Classification of Renal Cell Carcinoma on Biopsy and Surgical Resection Slides", Scientific Reports;11:7080 (2021).
title={Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides},
author={Zhu, Mengdan and Ren, Bing and Richards, Ryland and Suriawinata, Matthew and Tomita, Naofumi and Hassanpour, Saeed},
journal={Scientific reports},
volume={11},
number={1},
pages={1--9},
year={2021},
publisher={Nature Publishing Group}
}
FAQ
“I haven’t received any email after submitting the form.”
Please check your Junk/Spam email folder just in case the email got delivered there instead of your inbox.
If you still couldn’t find an email, please wait for a few hours and submit the form again.
“I received an email, but a download link has expired.”
By default, the download links will be expired after 4 hours. Please submit the form again to receive new links and download data before the links expire.
For inquiries, please contact us at ![]() BMIRDS.
BMIRDS.
If you are interested in histology image analysis, please check out other datasets from our group.




